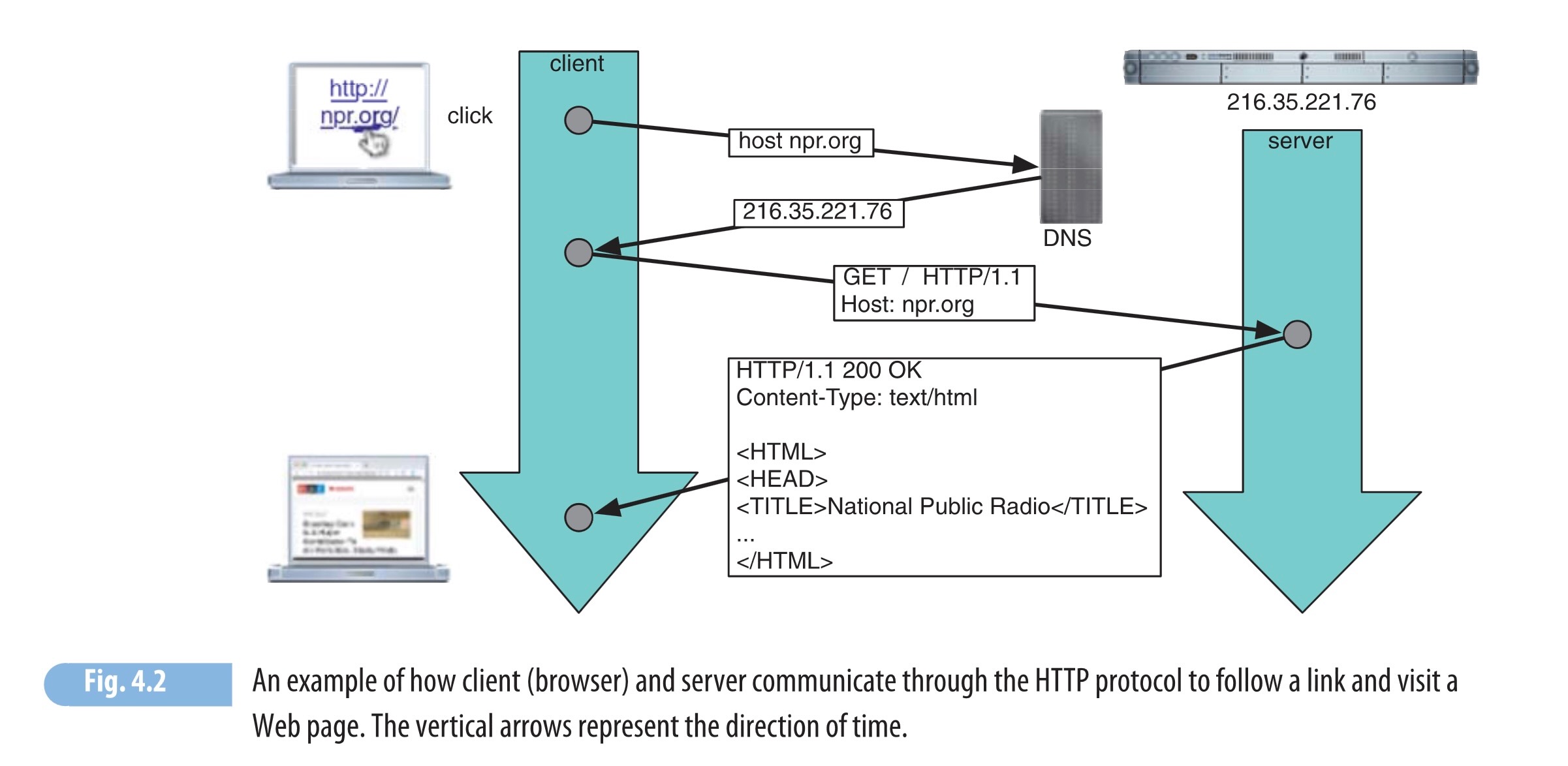
Week 4: Data Formats and APIs
DSAN 5000: Data Science and Analytics
Quiz Time
Data Gathering
- Preexisting data sources
- Web scraping
- Converting between formats
Preexisting Data Sources
- Depending on your field, or the type of data you’re looking for, there may be a “standard” data source! For example:
- Economics:
- US data: FRED
- Global data: World Bank Open Data, OECD Data, etc.
- Political Science:
- Network Science:
Web Scraping
- Fun fact: you can view a webpage’s HTML source code by right-clicking on the page and selecting “View Source”
- On older websites, this means we can just request page and parse the returned HTML
- Less fun fact: modern web frameworks (React, Next.js) generate pages dynamically using JS, meaning that what you see on the page will not be visible in the HTML source
- Data scraping still possible for these sites! Using browser automation tools like Selenium
Scraping Difficulty
| How is data loaded? | Solution | Example | ||
|---|---|---|---|---|
| 😊 | Easy | Data in HTML source | “View Source” | |
| 😐 | Medium | Data loaded dynamically via API | “View Source”, find API call, scrape programmatically | |
| 😳 | Hard | Data loaded dynamically [internally] via web framework | Use Selenium |
Data Structures: Foundations
Opening Datasets With Your Terminator Glasses On
- What does a row represent?
- What does a column represent?
- What does a value in a cell represent?
- Are there unique identifiers for the objects you care about?

From Raw Data to Clean Data
Data Structures: Simple \(\rightarrow\) Complex
| id | name | |
|---|---|---|
| 0 | K. Desbrow | kd9@dailymail.com |
| 1 | D. Minall | dminall1@wired.com |
| 2 | C. Knight | ck2@microsoft.com |
| 3 | M. McCaffrey | mccaf4@nhs.uk |
| year | month | points |
|---|---|---|
| 2023 | Jan | 65 |
| 2023 | Feb | |
| 2023 | Mar | 42 |
| 2023 | Apr | 11 |
| id | date | rating | num_rides |
|---|---|---|---|
| 0 | 2023-01 | 0.75 | 45 |
| 0 | 2023-02 | 0.89 | 63 |
| 0 | 2023-03 | 0.97 | 7 |
| 1 | 2023-06 | 0.07 | 10 |
| Source | Target | Weight |
|---|---|---|
| IGF2 | IGF1R | 1 |
| IGF1R | TP53 | 2 |
| TP53 | EGFR | 0.5 |
Tabular Data vs. Relational Data
- All of the datasets on the previous slide are tabular
- DBs like SQLite, MySQL model relationships within and between tabular datasets
- Imagine you’re creating the backend for a Georgetown social network. How would you record users and friendships? Your intuition may be record data:
| id | name | friends |
|---|---|---|
| 1 | Purna | [2,3,4] |
| 2 | Jeff | [1,3,4,5,6] |
| 3 | James | [1,2,4,6] |
| 4 | Britt | [1,2,3] |
| 5 | Dr. Fauci | [2,6] |
| 6 | Pitbull | [2,5] |
Long story short…
- This doesn’t scale
- Extremely inefficient to find whether two users are friends
- Redundant information: Have to store friendship between A and B in both A’s row and B’s row
A Better Approach
- Move the friendship data into its own table!
- This table now represents relational data, (user table still corresponds to records):
| user_id | name |
|---|---|
| 1 | Purna |
| 2 | Jeff |
| 3 | James |
| 4 | Britt |
| 5 | Dr. Fauci |
| 6 | Pitbull |
| id | friend_1 | friend_2 | id | friend_1 | friend_2 |
|---|---|---|---|---|---|
| 1 | 1 | 2 | 6 | 2 | 5 |
| 2 | 1 | 3 | 7 | 2 | 6 |
| 3 | 1 | 4 | 8 | 3 | 4 |
| 4 | 2 | 3 | 9 | 3 | 6 |
| 5 | 2 | 4 | 10 | 5 | 6 |
- May seem weird in terms of human readability, but think in terms of memory/computational efficiency: (a) Scalable, (b) Easy to find if two users are friends (via sorting/searching algorithms), (c) No redundant info
DBs: Relational or Otherwise
- For rest of lecture we zoom in on cases where data comes as individual files
- But on top of the relational format from previous slide, there are also non-relational database formats, like the document-based format used by e.g. MongoDB1
- In either case, data spread over many files \(\Rightarrow\) to obtain a single dataset we use queries.
Data Formats
- The most common formats, for most fields:
.csv: Comma-Separated Values.tsv: Tab-Separated Values.json: JavaScript Object Notation.xls/.xlsx: Excel format.dta: Stata format.yml: More human-readable alternative to JSON
.csv / .tsv
👍
my_data.csv
index,var_1,var_2,var_3
A,val_A1,val_A2,val_A3
B,val_B1,val_B2,val_B3
C,val_C1,val_C2,val_C3
D,val_D1,val_D2,val_D3(👎)
my_data.tsv
index var_1 var_2 var_3
A val_A1 val_A2 val_A3
B val_B1 val_B2 val_B3
C val_C1 val_C2 val_C3
D val_D1 val_D2 val_D3→
source("../_globals.r")
library(readr)
data <- read_csv("assets/my_data.csv")Rows: 3 Columns: 4
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (4): index, var_1, var_2, var_3
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.disp(data)# | index | var_1 | var_2 | var_3 |
# | - | - | - | - |
# | A | val_A1 | val_A2 | val_A3 |
# | B | val_B1 | val_B2 | val_B3 |
# | C | val_C1 | val_C2 | val_C3 |
# | D | val_D1 | val_D2 | val_D3 | - Python:
pd.read_csv()(from Pandas library) - R:
read_csv()(fromreadrlibrary)
.json
courses.json
{
"dsan5000": {
"title": "Data Science and Analytics",
"credits": 3,
"lectures": [
"Intro",
"Tools and Workflow"
]
},
"dsan5100": {
"title": "Probabilistic Modeling and Statistical Computing",
"credits": 3,
"lectures": [
"Intro",
"Conditional Probability"
]
}
}- Python:
json(built-in library,import json) - R:
jsonlite(install.packages("jsonlite")) - Helpful validator (for when
.jsonfile won’t load)
Other Formats
Appendix: Code Examples in R
Scraping HTML with httr2 and xml2
httr2 Documentation | xml2 Documentation
Code
library(httr2)
request_obj <- request("https://en.wikipedia.org/wiki/Data_science")
response_obj <- req_perform(request_obj)
# Parse HTML
library(xml2)
Attaching package: 'xml2'The following object is masked from 'package:httr2':
url_parseCode
html_obj <- response_obj |> resp_body_html()
html_obj |> xml_find_all('//h2') |> xml_text()[1] "Contents"
[2] "Foundations"
[3] "Etymology"
[4] "Data science and data analysis"
[5] "Data Science as an Academic Discipline"
[6] "Cloud computing for data science"
[7] "Ethical consideration in data science"
[8] "See also"
[9] "References" Note: httr2 is a re-written version of the original httr package, which is now deprecated. You’ll still see lots of code using httr, however, so it’s good to know how both versions work. Click here for a helpful vignette on the original httr library.
XPath I: Selecting Elements
mypage.html
<div class="container">
<h1>Header</h1>
<p id="page-content">Content</p>
<img class="footer-image m-5" src="footer.png">
</div>'//div'matches all elements<div>in the document:<div class="container"> <h1>Header</h1> <p id="page-content">Content</p> <img class="footer-image m-5" src="footer.png"> </div>'//div//img'matches<img>elements which are children of<div>elements:<img class="footer-image m-5" src="footer.png">
XPath II: Filtering by Attributes
mypage.html
<div class="container">
<h1>Header</h1>
<p id="page-content">Content</p>
<img class="footer-image m-5" src="footer.png">
</div>'//p[id="page-content"]'matches<p>elements with idpage-content3:<p id="page-content">Content</p>Matching classes is a bit trickier:
'//img[contains(concat(" ", normalize-space(@class), " "), " foot ")]'matches
<img>elements withfootas one of their classes4<img class="foot m-5" src="footer.png" />
Example: Math API
- Base URL:
https://newton.vercel.app/api/v2/ - The endpoint:
factor - The argument:
"x^2 - 1" - The request:
https://newton.vercel.app/api/v2/factor/x^2-1
Code
library(httr2)
request_obj <- request("https://newton.vercel.app/api/v2/factor/x^2-1")
response_obj <- req_perform(request_obj)
writeLines(response_obj %>% resp_body_string()){"operation":"factor","expression":"x^2-1","result":"(x - 1) (x + 1)"}Authentication
- Most APIs don’t allow requests to be made by anonymous requesters, and require authentication.
- For example, to access private GitHub repos using GitHub’s API, you’ll need to authenticate that you are in fact the one making the request
Authentication via GH
- The
GHlibrary forRcan handle this authentication process for you. For example, this private repo in my account does not show up if requested anonymously, but does show up if requested usingGHwith a Personal Access Token:
Code
library(gh)
tryCatch({
result <- gh("GET /repos/jpowerj/private-repo-test")
writeLines(paste0(result$name, ": ",result$description))
}, error = function(e) {
writeLines(paste0("Failed to authenticate: ",e))
})private-repo-test: Private repo example for DSAN5000- Your code should never contain authentication info, especially when using GitHub. In this case, I created an OS environment variable called
GITHUB_TOKENcontaining my Personal Access Token, whichGHthen uses to make authenticated requests - On Mac, this is accomplished by adding a line to your
~/.zprofilefile:export GITHUB_TOKEN="<token goes here>"
Footnotes
For (much) more on this topic, see this page from Prisma, a high-level “wrapper” that auto-syncs your DB structure with a TypeScript schema, so your code knows exactly “what’s inside” a variable whose content was retrieved from the DB…↩︎
Your code should 🚨never🚨 contain authentication info, especially when using GitHub. In this case, I created an OS environment variable called
GITHUB_TOKENcontaining my Personal Access Token, which I then loaded usingos.getenv()and provided toPyGithub.↩︎HTML requires
ids to be unique to elements (and elements cannot have more than oneid), meaning that this returns a single element for valid HTML code (not all webpages!). Also note the double-quotes afterid=, required in XPath↩︎Your intuition may be to just use
'//img[@class="footer-image"]'. Sadly, however, this will match only elements withfooter-imageas their only class. i.e., it will match<img class="footer-image">but not<img class="footer-image another-class">. This will usually fail, since most elements have several classes: e.g., in Bootstrap,<p class="p-5 m-3"></p>creates a<p>element with padding of 5 pixels and margin of 3 pixels.↩︎