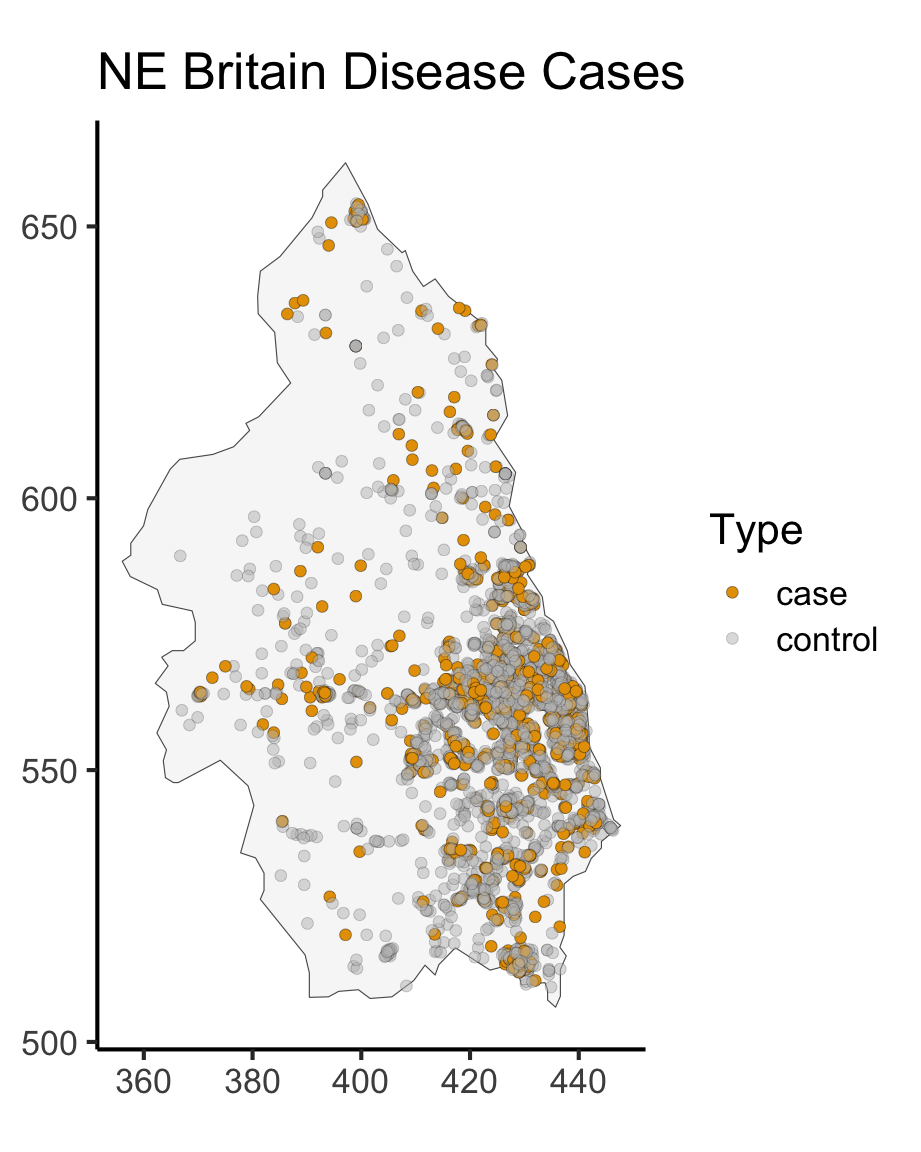
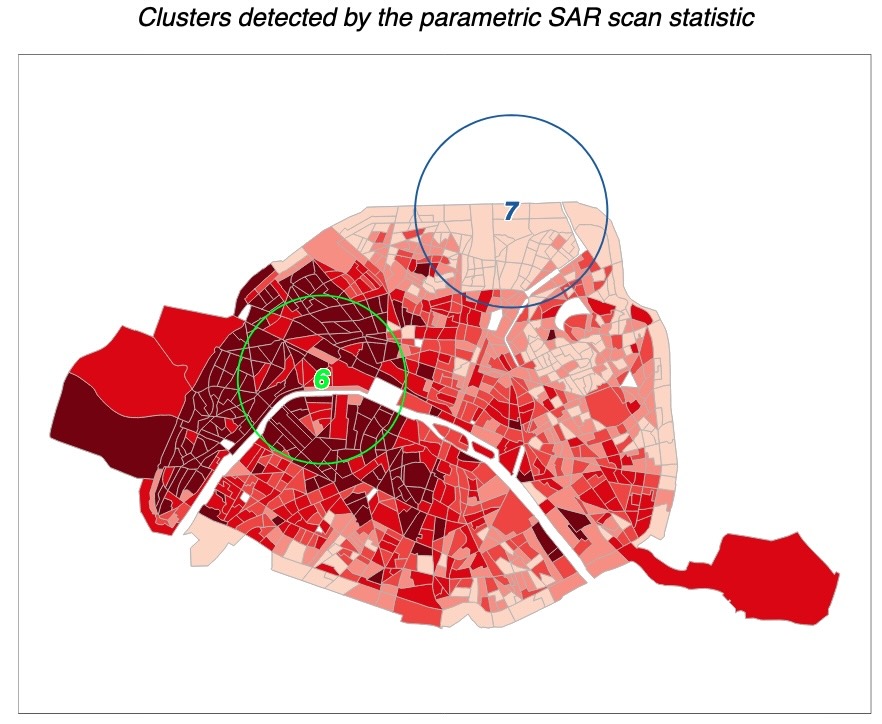
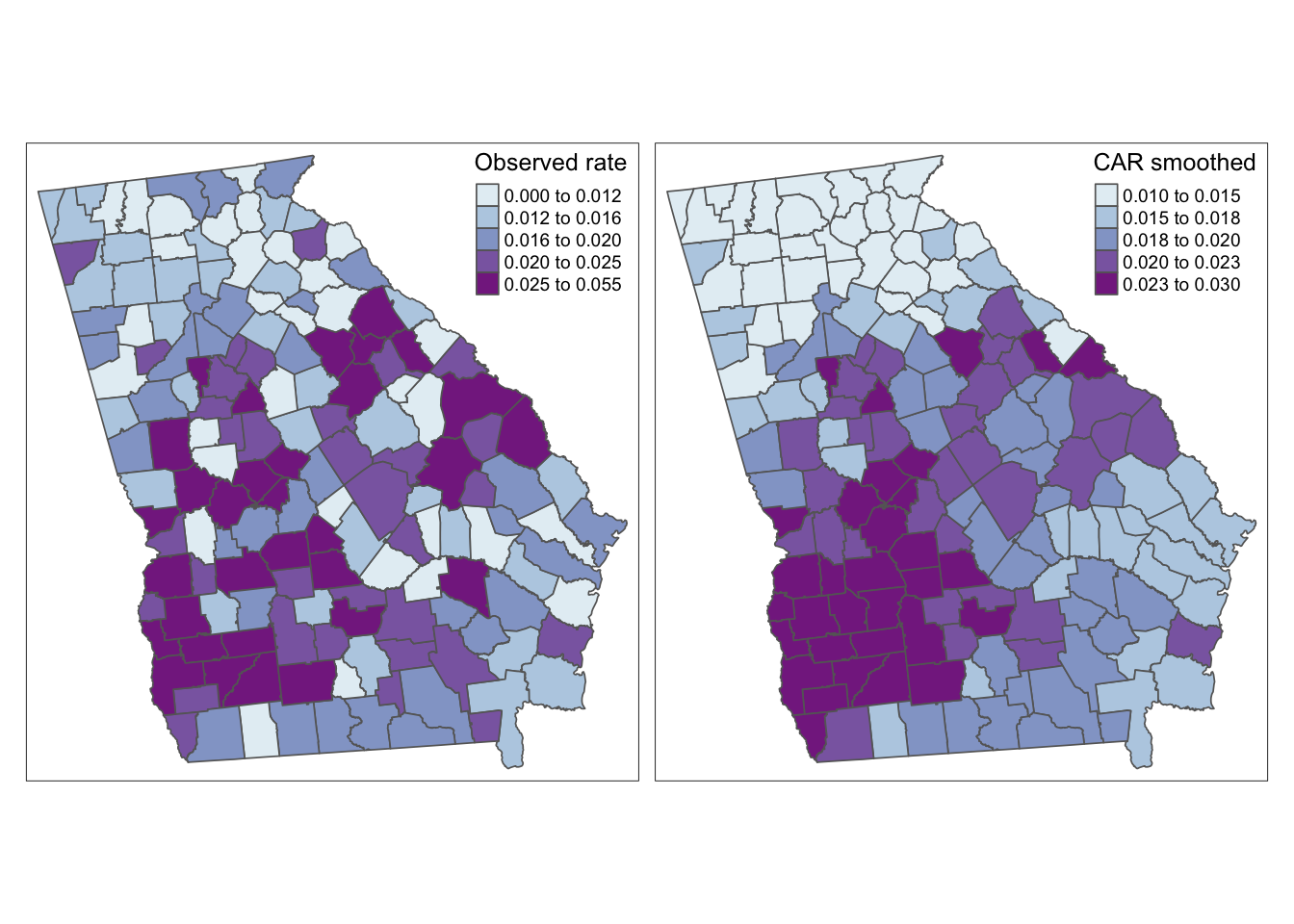
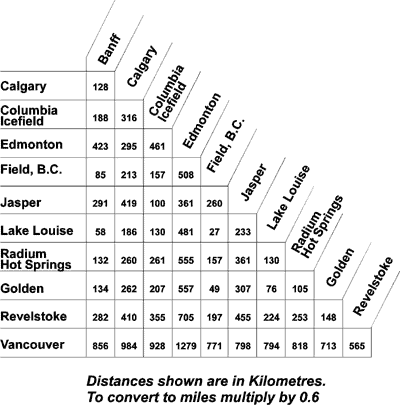
library(mapview) |> suppressPackageStartupMessages()
city_df <- tibble::tribble(
~city, ~lon, ~lat, ~pop,
"Chicago", 41.950567516553896, -87.93011127491978, 2746388,
"Detroit", 42.45123004999075, -83.18402319217698, 631524
)
city_sf <- sf::st_as_sf(
city_df,
coords = c("lat", "lon"),
crs=4326
)
city_buf_sf <- city_sf |> sf::st_buffer(20000)
city_cases_sf <- city_buf_sf |> sf::st_sample(size=rep(10,2)) |> sf::st_as_sf()
city_cases_sf$city <- "Detroit (10 Cases)"
city_cases_sf[1:10, 'city'] <- "Chicago (10 Cases)"
city_cases_sf$sample <- "Flu"
mapview(city_cases_sf, zcol="city")