Code
source("../dsan-globals/_globals.r")
set.seed(5300)DSAN 5300: Statistical Learning
Spring 2025, Georgetown University
Today’s Planned Schedule:
| Start | End | Topic | |
|---|---|---|---|
| Lecture | 6:30pm | 6:50pm | Roadmap: Week 4 \(\leadsto\) Week 6 → |
| 6:50pm | 7:20pm | Non-Linear Data-Generating Processes → | |
| 7:20pm | 8:00pm | Validation: Evaluating Non-Linear Models → | |
| Break! | 8:00pm | 8:10pm | |
| 8:10pm | 9:00pm | \(K\)-Fold Cross-Validation: Cooking with Gas → |
source("../dsan-globals/_globals.r")
set.seed(5300)\[ \DeclareMathOperator*{\argmax}{argmax} \DeclareMathOperator*{\argmin}{argmin} \newcommand{\bigexp}[1]{\exp\mkern-4mu\left[ #1 \right]} \newcommand{\bigexpect}[1]{\mathbb{E}\mkern-4mu \left[ #1 \right]} \newcommand{\definedas}{\overset{\small\text{def}}{=}} \newcommand{\definedalign}{\overset{\phantom{\text{defn}}}{=}} \newcommand{\eqeventual}{\overset{\text{eventually}}{=}} \newcommand{\Err}{\text{Err}} \newcommand{\expect}[1]{\mathbb{E}[#1]} \newcommand{\expectsq}[1]{\mathbb{E}^2[#1]} \newcommand{\fw}[1]{\texttt{#1}} \newcommand{\given}{\mid} \newcommand{\green}[1]{\color{green}{#1}} \newcommand{\heads}{\outcome{heads}} \newcommand{\iid}{\overset{\text{\small{iid}}}{\sim}} \newcommand{\lik}{\mathcal{L}} \newcommand{\loglik}{\ell} \DeclareMathOperator*{\maximize}{maximize} \DeclareMathOperator*{\minimize}{minimize} \newcommand{\mle}{\textsf{ML}} \newcommand{\nimplies}{\;\not\!\!\!\!\implies} \newcommand{\orange}[1]{\color{orange}{#1}} \newcommand{\outcome}[1]{\textsf{#1}} \newcommand{\param}[1]{{\color{purple} #1}} \newcommand{\pgsamplespace}{\{\green{1},\green{2},\green{3},\purp{4},\purp{5},\purp{6}\}} \newcommand{\prob}[1]{P\left( #1 \right)} \newcommand{\purp}[1]{\color{purple}{#1}} \newcommand{\sign}{\text{Sign}} \newcommand{\spacecap}{\; \cap \;} \newcommand{\spacewedge}{\; \wedge \;} \newcommand{\tails}{\outcome{tails}} \newcommand{\Var}[1]{\text{Var}[#1]} \newcommand{\bigVar}[1]{\text{Var}\mkern-4mu \left[ #1 \right]} \]
library(tidyverse) |> suppressPackageStartupMessages()
library(plotly) |> suppressPackageStartupMessages()
daly_df <- read_csv("assets/dalys_cleaned.csv")Rows: 162 Columns: 6
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (2): name, gdp_cap
dbl (4): dalys_pc, population, gdp_pc_clean, log_dalys_pc
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.daly_df <- daly_df |> mutate(
gdp_pc_1k=gdp_pc_clean/1000
)
model_labels <- labs(
x="GDP per capita ($1K PPP, 2021)",
y="Log(DALYs/n)",
title="Decrease in DALYs as GDP/n Increases"
)
daly_plot <- daly_df |> ggplot(aes(x=gdp_pc_1k, y=log_dalys_pc, label=name)) +
geom_point() +
# geom_smooth(method="loess", formula=y ~ x) +
geom_smooth(method="lm", formula=y ~ poly(x,5), se=FALSE) +
theme_dsan(base_size=14) +
model_labels
ggplotly(daly_plot)Warning: The following aesthetics were dropped during statistical transformation: label.
ℹ This can happen when ggplot fails to infer the correct grouping structure in
the data.
ℹ Did you forget to specify a `group` aesthetic or to convert a numerical
variable into a factor?\[ \begin{align*} \leadsto Y = &10.58 - 0.2346 X + 0.01396 X^2 \\ &- 0.0004 X^3 + 0.000005 X^4 \\ &- 0.00000002 X^5 + \varepsilon \end{align*} \]
eval_fitted_poly <- function(x) {
coefs <- c(
10.58, -0.2346, 0.01396,
-0.0004156, 0.0000053527, -0.0000000244
)
x_terms <- c(x^0, x^1, x^2, x^3, x^4, x^5)
dot_prod <- sum(coefs * x_terms)
return(dot_prod)
}
N <- 500
x_vals <- runif(N, min=0, max=90)
y_vals <- sapply(X=x_vals, FUN=eval_fitted_poly)
sim_df <- tibble(gdpc=x_vals, ldalys=y_vals)
ggplot() +
geom_line(data=sim_df, aes(x=gdpc, y=ldalys)) +
geom_point(data=daly_df, aes(x=gdp_pc_1k, y=log_dalys_pc)) +
theme_dsan() +
model_labels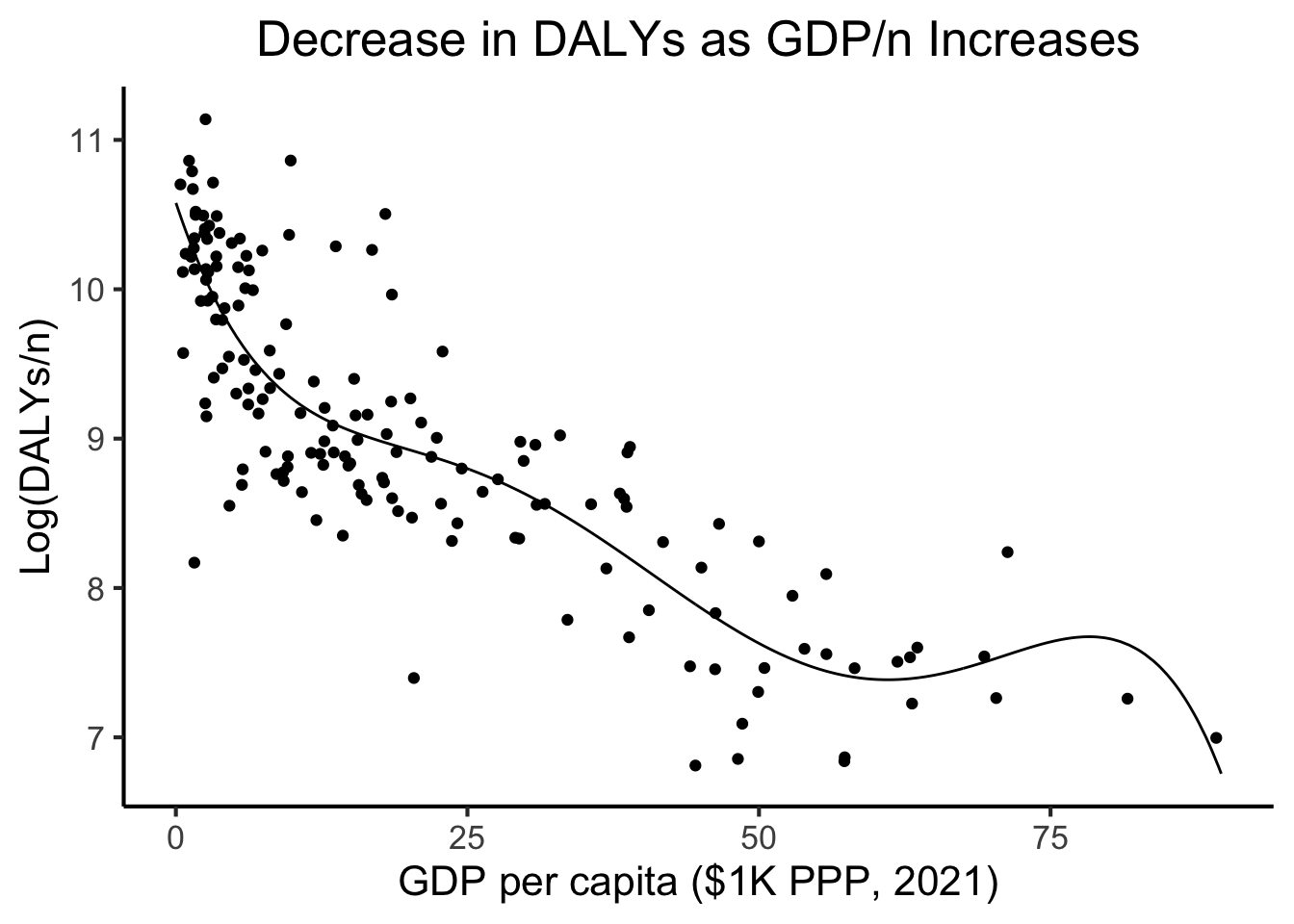
run_dgp <- function(world_label="Sim", N=60, x_max=90) {
x_vals <- runif(N, min=0, max=x_max)
y_raw <- sapply(X=x_vals, FUN=eval_fitted_poly)
y_noise <- rnorm(N, mean=0, sd=0.8)
y_vals <- y_raw + y_noise
sim_df <- tibble(
gdpc=x_vals,
ldalys=y_vals,
world=world_label
)
return(sim_df)
}
df1 <- run_dgp("World 1")
df2 <- run_dgp("World 2")
df3 <- run_dgp("World 3")
dgp_df <- bind_rows(df1, df2, df3)
dgp_df |> ggplot(aes(x=gdpc, y=ldalys)) +
geom_point(aes(color=world)) +
facet_wrap(vars(world)) +
theme_dsan(base_size=22) +
remove_legend() +
model_labels +
labs(title="Three Possible Realizations of our DGP")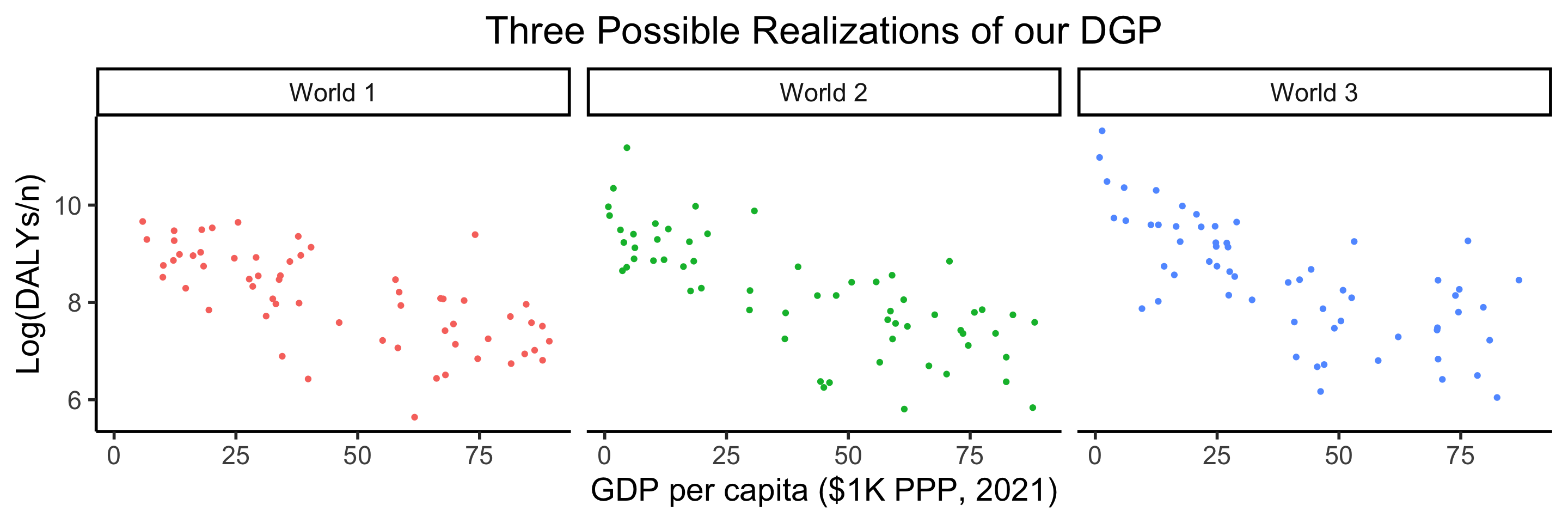
Specifically: evaluating non-linear models on how well they generalize!
graph grid
{
graph [
overlap=true,
scale=0.2
]
nodesep=0.0
ranksep=0.0
rankdir="LR"
scale=0.2
node [
style="filled",
color=black,
fillcolor=lightblue,
shape=box
]
// uncomment to hide the grid
edge [style=invis]
subgraph cluster_01 {
label="Training Set (80%)"
subgraph cluster_02 {
label="Training Fold (80%)"
A1[label="16%"] A2[label="16%"] A3[label="16%"] A4[label="16%"]
}
subgraph cluster_03 {
label="Validation Fold (20%)"
B1[label="16%",fillcolor=lightgreen]
}
}
subgraph cluster_04 {
label="Test Set (20%)"
C1[label="20%",fillcolor=orange]
}
A1 -- A2 -- A3 -- A4 -- B1 -- C1;
}library(boot)
set.seed(5300)
sim200_df <- run_dgp(
world_label="N=200", N=200, x_max=100
)
sim1k_df <- run_dgp(
world_label="N=1000", N=1000, x_max=100
)
compute_deltas <- function(df, min_deg=1, max_deg=12) {
cv_deltas <- c()
for (i in min_deg:max_deg) {
cur_poly <- glm(ldalys ~ poly(gdpc, i), data=df)
cur_poly_cv_result <- cv.glm(data=df, glmfit=cur_poly, K=5)
cur_cv_adj <- cur_poly_cv_result$delta[1]
cv_deltas <- c(cv_deltas, cur_cv_adj)
}
return(cv_deltas)
}
sim200_deltas <- compute_deltas(sim200_df)
sim200_delta_df <- tibble(degree=1:12, delta=sim200_deltas)
sim200_delta_df |> ggplot(aes(x=degree, y=delta)) +
geom_line() +
geom_point() +
geom_vline(xintercept=5, linetype="dashed") +
scale_x_continuous(
breaks=seq(from=1,to=12,by=1)
) +
theme_dsan(base_size=22) +
labs(title="N = 200")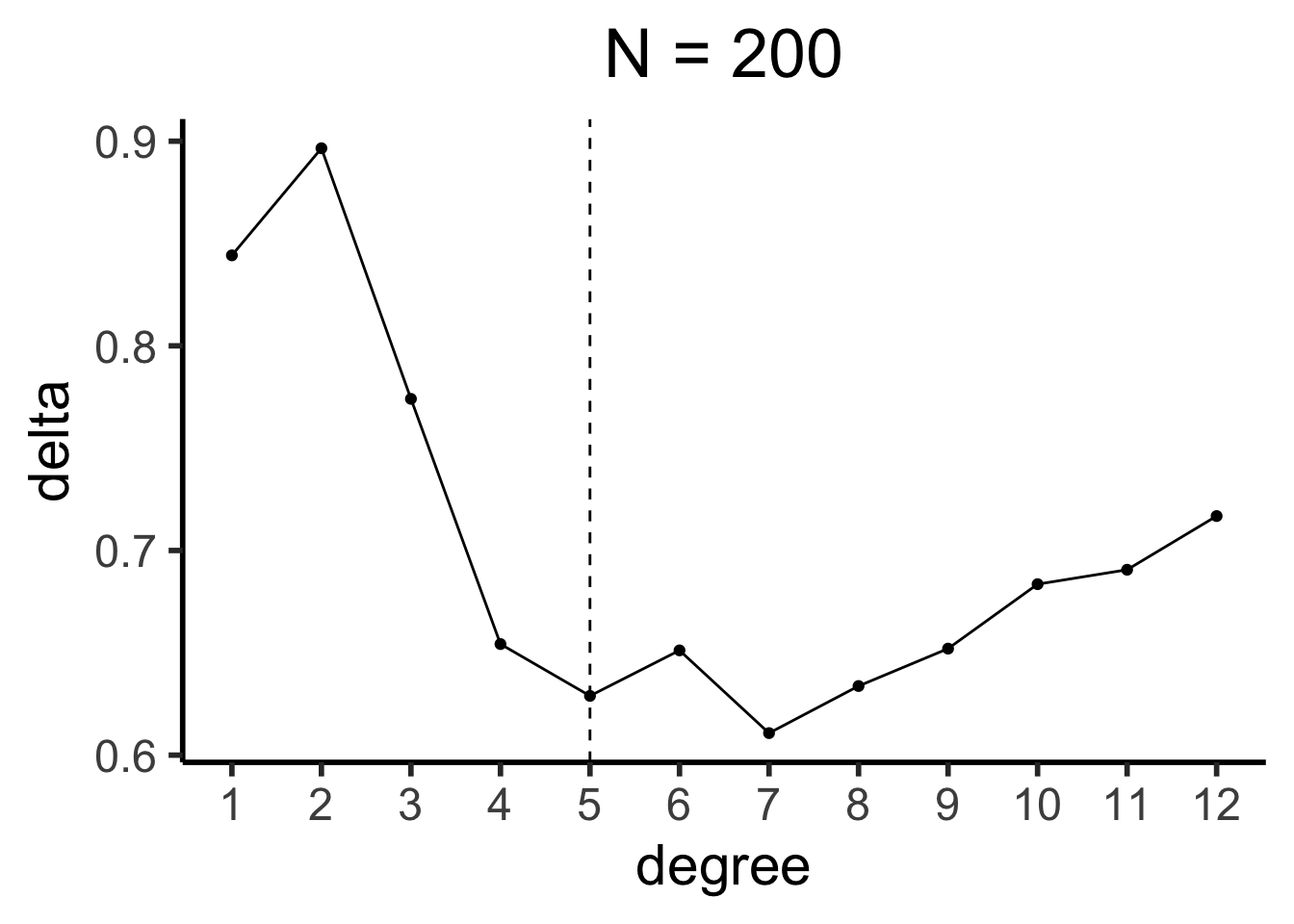
sim1k_deltas <- compute_deltas(sim1k_df)
sim1k_delta_df <- tibble(degree=1:12, delta=sim1k_deltas)
sim1k_delta_df |> ggplot(aes(x=degree, y=delta)) +
geom_line() +
geom_point() +
geom_vline(xintercept=5, linetype="dashed") +
scale_x_continuous(
breaks=seq(from=1,to=12,by=1)
) +
theme_dsan(base_size=22) +
labs(title="N = 1000")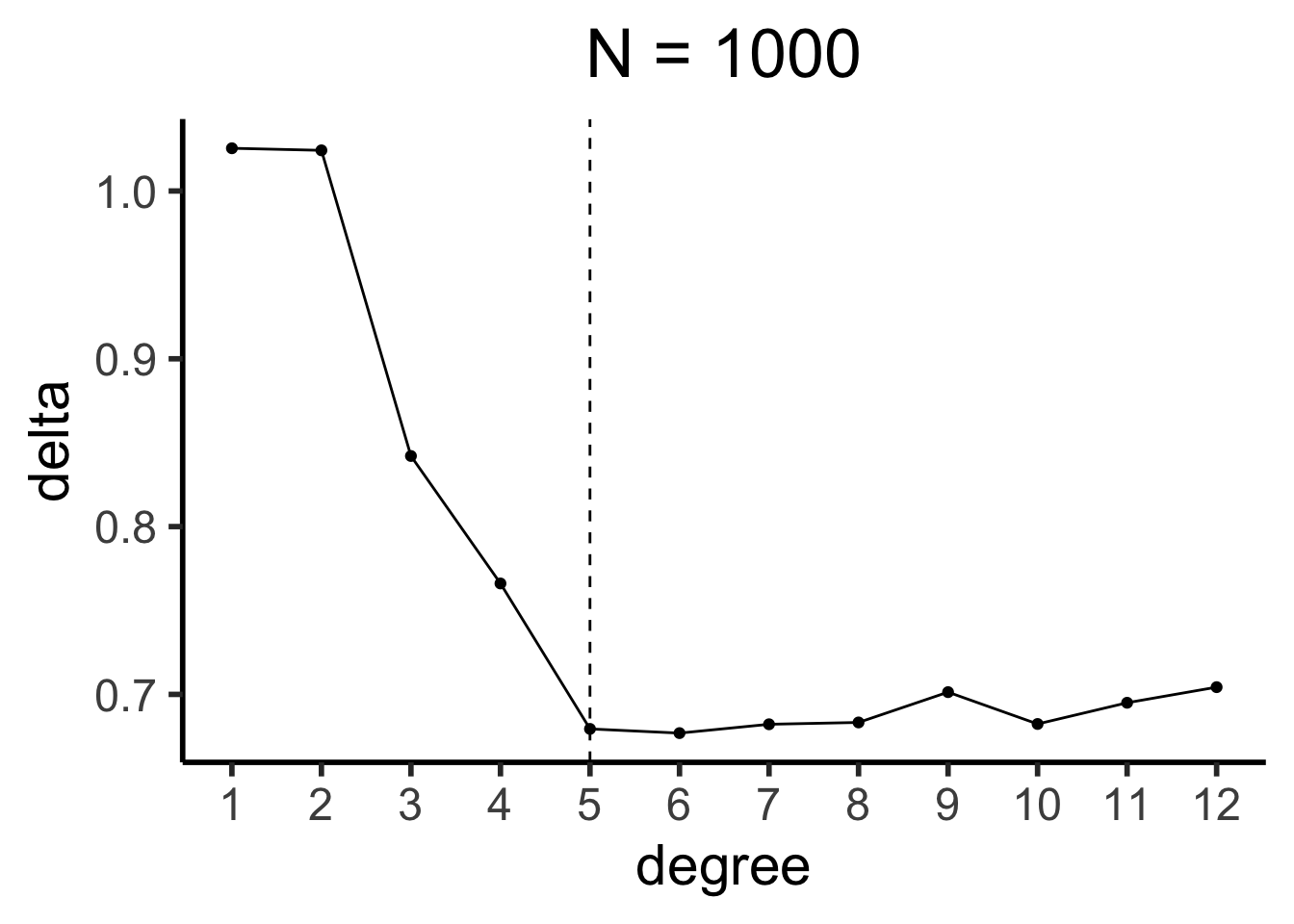
(加 even more 油!)
\[ \varepsilon_{(K)} = \frac{1}{K}\sum_{i=1}^{K}\varepsilon^{\text{Val}}_{i} \]
\[ \begin{align*} \varepsilon_{(N)} =~ & \text{Err}(\mathbf{D}_1 \mid \mathbf{D}_{2:100}) + \text{Err}(\mathbf{D}_2 \mid \mathbf{D}_1, \mathbf{D}_{3:100}) \\ &+ \text{Err}(\mathbf{D}_3 \mid \mathbf{D}_{1:2}, \mathbf{D}_{4:100}) + \cdots + \text{Err}(\mathbf{D}_{100} \mid \mathbf{D}_{1:99}) \end{align*} \]